

In my research, I mainly ask how macroevolutionary processes have formed the patterns of biodiversity we observe today.
My tool of choice to address this question is phylogenetic comparative methods (PCM), with a main focus is on lineage diversification, but also trait evolution, biogeography, and their interplay. My main research interests cover three different aspects:
Past Research: More on past projects from my BSc, MSc, and PhD.
Software & Code: More on packages & snippets related to the work below.
Beyond finding our way among the various different methods and tools we have at our disposal to analyse adaptive radiations, weighing their strengths and limitations, a main challenge for macroevolutionary biologists is to be able to tell what biological interpretations our results even allow for. For example, while trait-dependent diversification (SSE) models let us find associations between trait states and diversification rates, their results are mainly correlational, and our ability to confidently make inferences on mechanisms or causal relations is limited.
I thus, in the spirit of Phylogenetic Natural History (PNH) I aim to combine data-driven and hypothesis-testing approaches, and make use of graphical models and causal inference for phylogenetics. I make use of DAGs (Directed Acyclic Graphs) to depict and test concrete causal hypotheses, and incorporate approaches like path analysis to test competing hypotheses against each other.
The goal is to be better able to distinguish alternative causal scenarios, by explicitly testing conditional dependencies between parameters; to better incorporate confounding latent factors (e.g. singular events that affected certain clades); and to formulate and test more realistic hypotheses on adaptive radiations, by accounting for ecological opportunities.
This approach will allow for much more nuanced results, e.g., establishing which radiations can and which cannot be explained by our hypotheses, and assessing the relative importance of a particular factor in explaining diversification rate shifts. Furthermore, this approach will in the long run allow for us as a field to have more clarity on hypotheses, and to more easily build upon previous findings (by carrying forth and expanding the DAGs).
I am currently pursuing this work in the Uyeda Lab at Virginia Tech, and other collaborators.
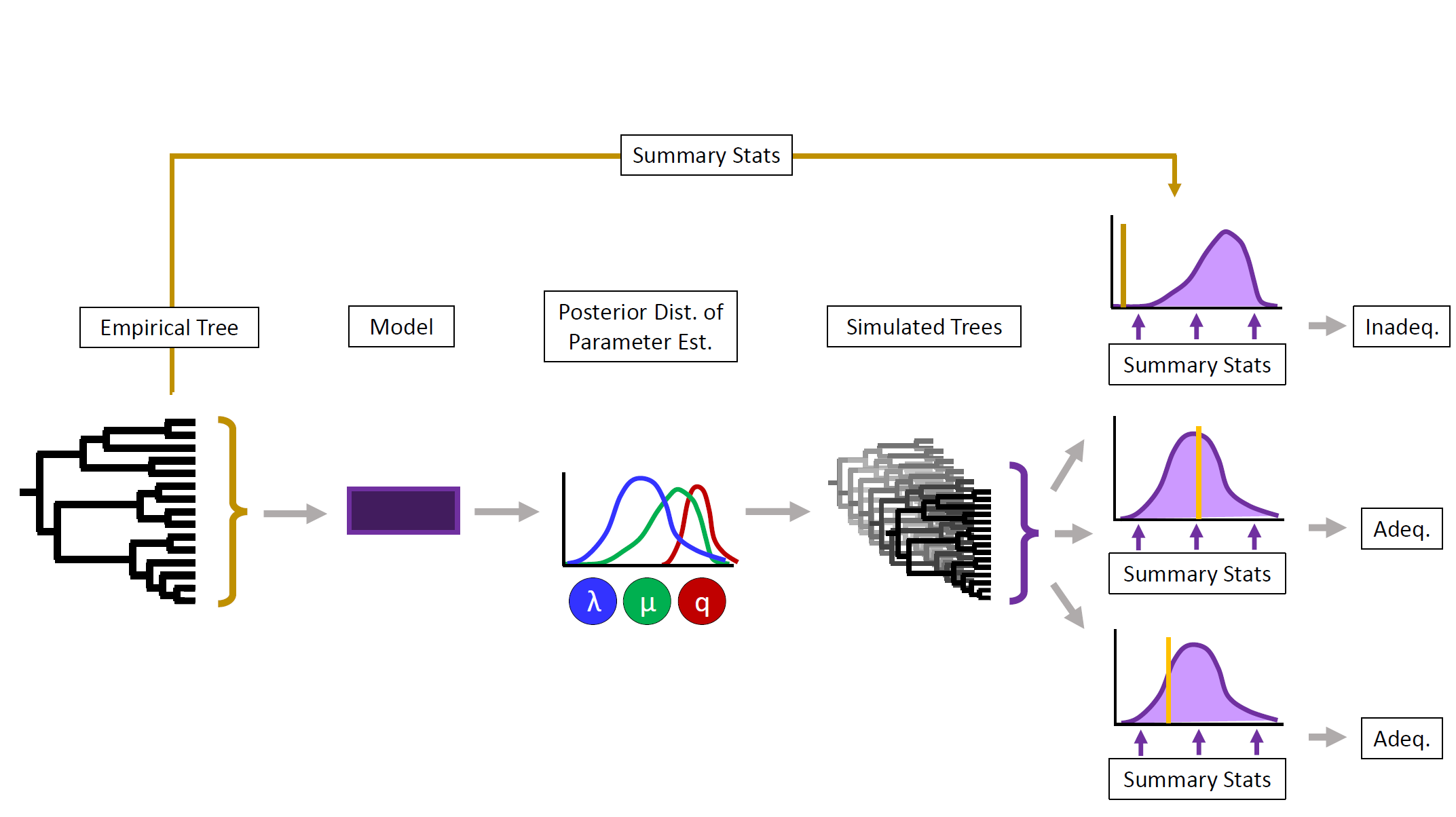
To study diversification, we have a large number of different models at our disposal, covering a range of complexity, and incorporating a variety of different factors and processes. However, all of them suffer from known and unknown limitations, biases, and violated assumptions, casting doubt on our results. Traditional model testing via likelihoods or AIC merely allows to rank models by their fit, but allows no insight into whether their inferences can be trusted in the light of our data.
One way to address this, is to ask whether a model is able to accurately describe our data to begin with. This can be done using model adequacy tests. The underlying logic is, that for a model to be adequate for our data, it should generate data that ‘looks similar’ to ours when simulated under the same parameters. Hence, discrepancies between the original data and the simulations would let us quantify the shortcomings of a model to describe the data adequately, and explore processes that are still missing from it. Beyond model selection, adequacy tests could have a positive impact on the choice of reasonable null models, model averaging, as well as development and validation of new approaches.
So far, this line of work as resulted in two approaches: The R package BoskR tests adequacy for simple birth death models (Yule, BD, time-, and density-dependent BD) and more in a Maximum Likelihood (ML) framework, and which is descriped in this preprint. The procedure adequaSSE in RevBayes tests adequacy for trait-dependent diversification (SSE) models in a Bayesian framework using posterior predictive simulations (PPS). An early manuscript is available as a preprint. Using these, we were able to sucessfully identify false inferences of trait dependence. We chould further demonstrate the potential to distinguish the nature and extent of the data’s departure from the model assumptions.
This work has been done together with Brian O’Meara at UTK, Emma Goldberg at the NMC, and Will Freyman at 23andMe.

Starting out as a botanist, and later working on insects, I found that I am more fascinated with answering macroevolutionary questions than that I am married to a specific group of organisms. And the fact that my work is driven by methods development and computational analyses opens many opportunities for me to collaborate. I am thus always happy to broaden my horizon by applying my skills to questions in a new taxonomic group.
My empirical interests usually focus on differences in diversification among lineages and the factors that influenced it. This often involves testing trait-dependent diversification, trait evolution, or biogeography.

Examples of empirical projects I conducted or was involved with include: